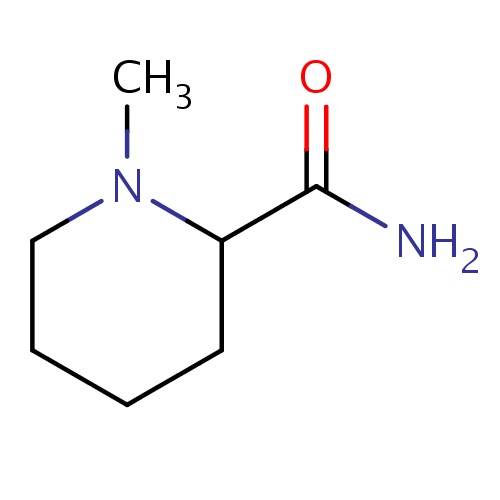
Common name
(2S)-1-methylpiperidine-2-carboxamide
IUPAC name
(2S)-1-methylpiperidine-2-carboxamide
SMILES
CN1C(CCCC1)C(=O)N
Common name
(2S)-1-methylpiperidine-2-carboxamide
IUPAC name
(2S)-1-methylpiperidine-2-carboxamide
SMILES
CN1C(CCCC1)C(=O)N
INCHI
InChI=1S/C7H14N2O/c1-9-5-3-2-4-6(9)7(8)10/h6H,2-5H2,1H3,(H2,8,10)/t6-/m0/s1
FORMULA
C7H14N2O

Common name
(2S)-1-methylpiperidine-2-carboxamide
IUPAC name
(2S)-1-methylpiperidine-2-carboxamide
Molecular weight
142.199
clogP
0.069
clogS
-0.326
Frequency
0.0010
HBond Acceptor
2
HBond Donor
2
Total PolarSurface Area
46.33
Number of Rings
1
Rotatable Bond
1
| Drug ID | Common name | Structure CAS | Compound class | Therapeutic area |
|---|---|---|---|---|
| FDBD00183 | Ropivacaine |
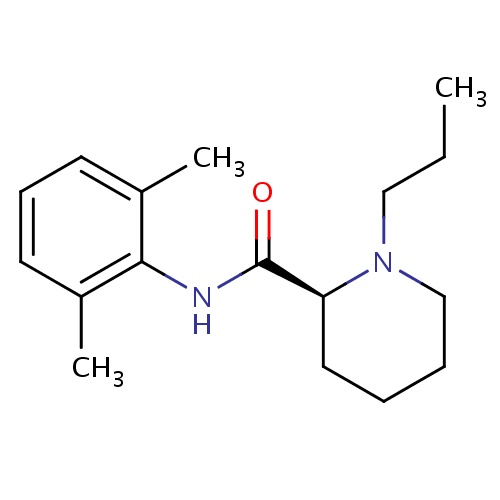
|
Anesthetics, Local; Anesthetics; Nervous System; Amides; Cytochrome P-450 CYP1A2 Inhibitors; Cytochrome P-450 CYP1A2 Inducers; Cytochrome P-450 CYP2B6 Inducers; Cytochrome P-450 CYP2B6 Inhibitors; CYP2B6 Inhibitors (strong); CYP2D6 Inducers; CYP2D6 Inducers (strong); CYP3A4 Inhibitors; | Used in obstetric anesthesia and regional anesthesia for surgery. |
| FDBD00184 | Bupivacaine |
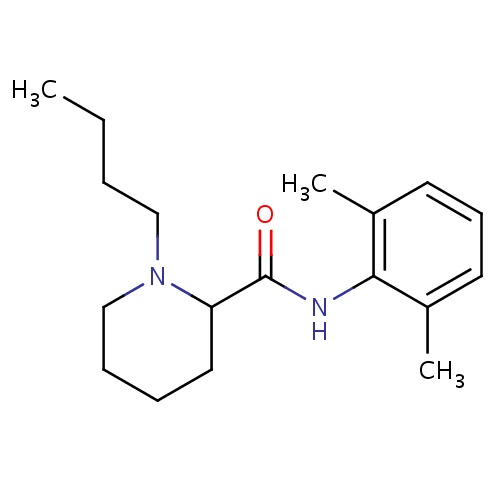
|
Anesthetics, Local; Anesthetics; Nervous System; Amides; Cytochrome P-450 CYP1A2 Inhibitors; Cytochrome P-450 CYP1A2 Inducers; Cytochrome P-450 CYP2C19 Inducers; CYP2D6 Inducers; CYP2D6 Inducers (strong); CYP3A4 Inhibitors; | For the production of local or regional anesthesia or analgesia for surgery, for oral surgery procedures, for diagnostic and therapeutic procedures, and for obstetrical procedures. |
| FDBD00856 | Levobupivacaine |
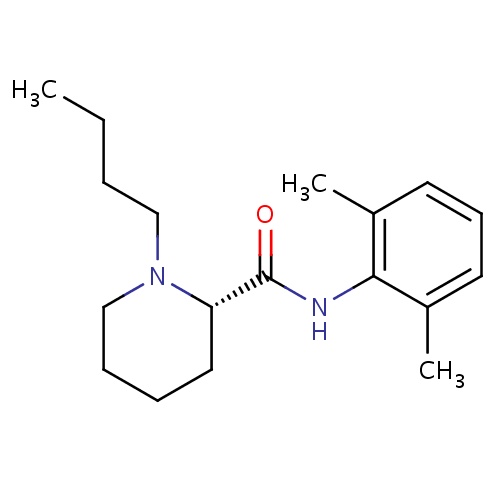
|
Anesthetics, Local; Anesthetics; Nervous System; Amides; Cytochrome P-450 CYP1A2 Inhibitors; Cytochrome P-450 CYP1A2 Inducers; CYP3A4 Inhibitors; | For the production of local or regional anesthesia for surgery and obstetrics, and for post-operative pain management. |
3 ,
1
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
| 1b6l_ligand_2_15.mol2 | 1b6l | 1 | -6.19 | C(=O)(N)[C@H]1[N@@H+](CCCC1)C | 10 |
| 1idb_ligand_2_46.mol2 | 1idb | 1 | -6.08 | C[N@H+]1[C@@H](CCCC1)C(=O)N | 10 |
| 1ida_ligand_2_64.mol2 | 1ida | 1 | -6.02 | C(=O)(N)[C@H]1[N@@H+](CCCC1)C | 10 |
| 3cyx_ligand_2_75.mol2 | 3cyx | 0.921053 | -7.04 | C[N@H+]1[C@@H](C[C@@H]2CCCC[C@@H]2C1)C(=O)N | 14 |
| 3ekq_ligand_2_75.mol2 | 3ekq | 0.921053 | -7.02 | C(=O)(N)[C@H]1[N@@H+](C[C@@H]2[C@H](C1)CCCC2)C | 14 |
| 4qgi_ligand_2_63.mol2 | 4qgi | 0.921053 | -7.02 | [N@@H+]1([C@@H](C[C@@H]2CCCC[C@@H]2C1)C(=O)N)C | 14 |
| 2pyn_ligand_2_0.mol2 | 2pyn | 0.921053 | -7.00 | C[N@H+]1[C@H](C(=O)N)C[C@H]2[C@H](CCCC2)C1 | 14 |
| 3ekx_ligand_2_0.mol2 | 3ekx | 0.921053 | -7.00 | C(=O)(N)[C@@H]1C[C@H]2[C@H](CCCC2)C[N@H+]1C | 14 |
104 ,
11

