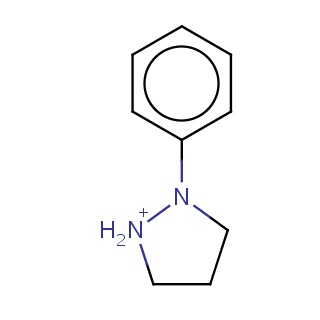
Binding information for 3fi2_ligand_1_5.mol2(FDBF05055)
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
| 3fi2_ligand_1_5.mol2 | 3fi2 | 0.5 | -6.94 | c1cccc(c1)N1CCC[NH2+]1 | 11 |
Structure and binding mode of 3fi2_ligand_1_5.mol2(FDBF05055)
Important binding residues for 3fi2_ligand_1_5.mol2(FDBF05055)
| PDBID | RESIDUE | ΔEvdw | ΔEele | ΔEgas | ΔEsolv | ΔEgb |
|---|---|---|---|---|---|---|
| 3fi2 | ILE70 | -1.78 | -17.65 | -19.43 | 17.77 | -1.66 |
| 3fi2 | MET146 | -0.89 | -0.61 | -1.5 | 0.65 | -0.85 |
| 3fi2 | GLU147 | -0.38 | -22.67 | -23.05 | 21.16 | -1.88 |
| 3fi2 | ASP150 | -1.03 | -21.87 | -22.9 | 21.38 | -1.52 |
| 3fi2 | ALA151 | -1.15 | -0.40 | -1.55 | 0.54 | -1.00 |
| 3fi2 | ASN152 | -0.88 | 1.76 | 0.88 | -1.39 | -0.51 |
| 3fi2 | VAL197 | -0.15 | -1.46 | -1.61 | 1.20 | -0.40 |
| 3fi2 | LEU206 | -0.65 | 0.34 | -0.31 | -0.14 | -0.45 |