Binding information for 1l6s_ligand_frag_0.mol2(FDBF00004)
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
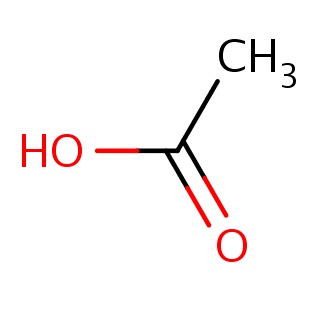
| 1l6s_ligand_frag_0.mol2 | 1l6s | 1 | -5.92 | CC(=O)O | 4 |
Structure and binding mode of 1l6s_ligand_frag_0.mol2(FDBF00004)
Important binding residues for 1l6s_ligand_frag_0.mol2(FDBF00004)
| PDBID | RESIDUE | ΔEvdw | ΔEele | ΔEgas | ΔEsolv | ΔEgb |
|---|---|---|---|---|---|---|
| 1l6s | TYR200 | -0.81 | -0.75 | -1.56 | -0.65 | -2.21 |
| 1l6s | PRO202 | -0.11 | -1.69 | -1.8 | 1.33 | -0.47 |
| 1l6s | PHE203 | -0.22 | -2.82 | -3.04 | 2.26 | -0.78 |
| 1l6s | TYR269 | -0.71 | -20.10 | -20.81 | 19.93 | -0.88 |
| 1l6s | GLN270 | -0.09 | -0.17 | -0.26 | -0.21 | -0.47 |
| 1l6s | VAL271 | -0.40 | -7.87 | -8.27 | 2.17 | -6.09 |
| 1l6s | SER272 | 2.02 | -19.95 | -17.93 | 11.86 | -6.07 |