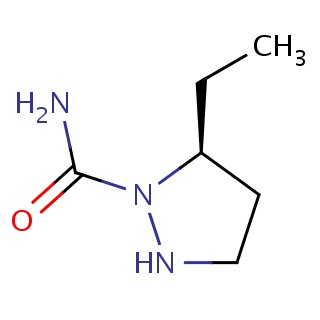
Binding information for 2g1q_ligand_2_10.mol2(FDBF02647)
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
| 2g1q_ligand_2_10.mol2 | 2g1q | 0.525424 | -6.02 | C1NN([C@H](C1)CC)C(=O)N | 10 |
Structure and binding mode of 2g1q_ligand_2_10.mol2(FDBF02647)
Important binding residues for 2g1q_ligand_2_10.mol2(FDBF02647)
| PDBID | RESIDUE | ΔEvdw | ΔEele | ΔEgas | ΔEsolv | ΔEgb |
|---|---|---|---|---|---|---|
| 2g1q | GLU118 | -0.61 | 0.16 | -0.45 | 0.11 | -0.34 |
| 2g1q | ARG119 | -0.98 | -5.10 | -6.08 | 4.67 | -1.41 |
| 2g1q | PRO137 | -0.45 | 0.08 | -0.37 | -0.02 | -0.39 |
| 2g1q | TYR211 | -0.88 | 0.05 | -0.83 | 0.36 | -0.47 |
| 2g1q | LEU214 | -1.27 | -0.78 | -2.05 | 0.64 | -1.41 |
| 2g1q | ALA218 | -0.51 | 0.34 | -0.17 | -0.47 | -0.64 |