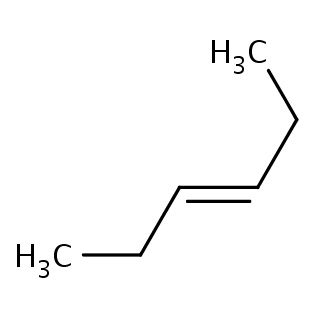
Binding information for 5d1r_ligand_5_246.mol2(FDBF00116)
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
| 5d1r_ligand_5_246.mol2 | 5d1r | 1 | -6.54 | C(=C\CC)/CC | 6 |
Structure and binding mode of 5d1r_ligand_5_246.mol2(FDBF00116)
Important binding residues for 5d1r_ligand_5_246.mol2(FDBF00116)
| PDBID | RESIDUE | ΔEvdw | ΔEele | ΔEgas | ΔEsolv | ΔEgb |
|---|---|---|---|---|---|---|
| 5d1r | VAL139 | -1.12 | -0.10 | -1.22 | -0.12 | -1.35 |
| 5d1r | PHE143 | -0.55 | -0.01 | -0.56 | 0.23 | -0.32 |
| 5d1r | PHE190 | -1.04 | -0.08 | -1.12 | 0.52 | -0.59 |
| 5d1r | TRP193 | -0.96 | 0.19 | -0.77 | 0.29 | -0.48 |
| 5d1r | TYR208 | -1.17 | 0.01 | -1.16 | 0.65 | -0.51 |
| 5d1r | MET212 | -0.77 | 0.21 | -0.56 | 0.20 | -0.36 |