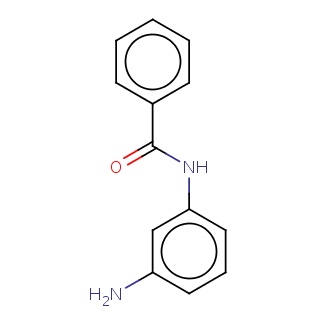
Binding information for 1y57_ligand_3_9.mol2(FDBF03278)
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
| 1y57_ligand_3_9.mol2 | 1y57 | 0.666667 | -7.02 | O=C(Nc1cccc(c1)N)c1ccccc1 | 16 |
Structure and binding mode of 1y57_ligand_3_9.mol2(FDBF03278)
Important binding residues for 1y57_ligand_3_9.mol2(FDBF03278)
| PDBID | RESIDUE | ΔEvdw | ΔEele | ΔEgas | ΔEsolv | ΔEgb |
|---|---|---|---|---|---|---|
| 1y57 | LEU273 | -2.68 | 0.65 | -2.03 | -0.29 | -2.33 |
| 1y57 | GLY274 | -0.93 | -0.12 | -1.05 | 0.16 | -0.89 |
| 1y57 | VAL281 | -0.76 | 0.24 | -0.52 | -0.36 | -0.88 |
| 1y57 | TYR340 | -1.09 | -0.80 | -1.89 | 1.51 | -0.38 |
| 1y57 | MET341 | -0.05 | -2.47 | -2.52 | 1.51 | -1.01 |
| 1y57 | LYS343 | -0.42 | -0.13 | -0.55 | -0.00 | -0.55 |
| 1y57 | GLY344 | -1.10 | -0.98 | -2.08 | 0.74 | -1.34 |
| 1y57 | LEU393 | -0.61 | 0.09 | -0.52 | -0.14 | -0.65 |