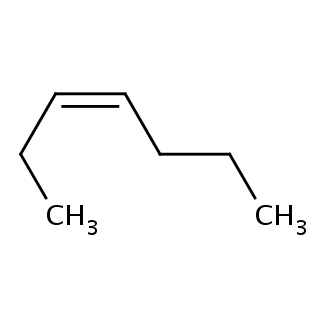
Binding information for 1gnj_ligand_4_700.mol2(FDBF00118)
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
| 1gnj_ligand_4_700.mol2 | 1gnj | 1 | -6.76 | CCC/C=C\CC | 7 |
Structure and binding mode of 1gnj_ligand_4_700.mol2(FDBF00118)
Important binding residues for 1gnj_ligand_4_700.mol2(FDBF00118)
| PDBID | RESIDUE | ΔEvdw | ΔEele | ΔEgas | ΔEsolv | ΔEgb |
|---|---|---|---|---|---|---|
| 1gnj | TYR138 | -0.67 | 0.05 | -0.62 | 0.04 | -0.58 |
| 1gnj | LEU139 | -1.09 | 0.04 | -1.05 | -0.02 | -1.07 |
| 1gnj | ILE142 | -1.31 | -0.11 | -1.42 | -0.23 | -1.64 |
| 1gnj | LEU154 | -0.92 | -0.18 | -1.1 | 0.37 | -0.73 |
| 1gnj | PHE157 | -1.24 | -0.02 | -1.26 | 0.09 | -1.17 |
| 1gnj | ALA158 | -0.92 | 0.09 | -0.83 | -0.05 | -0.88 |
| 1gnj | TYR161 | -1.03 | -0.14 | -1.17 | 0.54 | -0.63 |