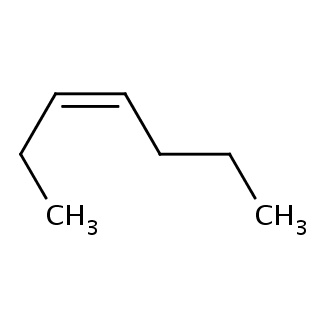
Binding information for 1gnj_ligand_4_220.mol2(FDBF00118)
| FRAGNAME | PDBID | SIMILIRITY | XSCORE | SMILE | HAC |
|---|---|---|---|---|---|
| 1gnj_ligand_4_220.mol2 | 1gnj | 1 | -6.66 | C(CC)/C=C\CC | 7 |
Structure and binding mode of 1gnj_ligand_4_220.mol2(FDBF00118)
Important binding residues for 1gnj_ligand_4_220.mol2(FDBF00118)
| PDBID | RESIDUE | ΔEvdw | ΔEele | ΔEgas | ΔEsolv | ΔEgb |
|---|---|---|---|---|---|---|
| 1gnj | LEU135 | -0.80 | 0.00 | -0.8 | 0.15 | -0.64 |
| 1gnj | TYR138 | -1.50 | 0.14 | -1.36 | 0.47 | -0.89 |
| 1gnj | ILE142 | -0.25 | -0.00 | -0.25 | -0.08 | -0.33 |
| 1gnj | TYR161 | -1.18 | -0.08 | -1.26 | 0.55 | -0.71 |
| 1gnj | PHE165 | -0.67 | -0.04 | -0.71 | 0.39 | -0.32 |
| 1gnj | LEU182 | -0.46 | -0.02 | -0.48 | 0.04 | -0.43 |